Insights of Epigenetics and Chromatin
Distinct genetic variants of early and late-onset prostate cancer
Victor Chalfant#, Samuel Serrano#, Mohamed Al-Toubat*, Carlos Riveros#, Ahmed Elshafei and KC Balaji
#These authors contributed equally to this work
Cite this as
Chalfant V, Serrano S, Al-Toubat M, Riveros C, Elshafei A, et al. (2023) Distinct genetic variants of early and late-onset prostate cancer. Insights Epigenet Chromatin. 1(1): 001-008. DOI: 10.17352/iec.000001Copyright Licence
© 2023 Chalfant V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Background: The incidence of early-onset Prostate Cancer (PCa) has increased in the last two decades. Men diagnosed with PCa before age 55 have lower 5-year relative survival rates compared to patients diagnosed later in life. Given the enhanced lethality of early-onset PCa, our aim is to evaluate somatic differences between early and late-onset PCa.
Methods: Patients with PCa were dichotomized into early (< 55 years old) and late-onset PCa (≥ 55 years old). Data is derived from the American Association for Cancer Research Project Genomics Evidence Neoplasia Information Exchange (GENIE) registry. The GENIE registry contains sequenced tumor samples and clinical data across many cancers a total of 4,546 Patients and 5,740 samples were included with pathologically confirmed prostate adenocarcinoma. The data is derived from 17 cancer centers from 2011 to 2021. Patterns in somatic gene tumor profiles were compared between early-onset and late-onset PCa using a chi-square test and logistic regression.
Results: A total of 452 (11.0%) patients had early-onset PCa while 3640 (89.0%) patients had late-onset PCa. Patients with early-onset PCa were more likely to be Black (12.2% vs. 7.7%) and less likely to have metastatic disease (32.0% vs. 45.0%). After logistic regression, early-onset PCa patients had higher odds of having a mutation in CDK12 [1.51 (95% CI: 1.04-2.22)] and ERF [1.81 (95% CI: 1.02-3.24)]. Patients with a CDK12 mutation were more likely to be Black [1.92 (95% CI: 1.28-2.86); P = 0.002) and to have metastatic disease [1.53 (95% CI: 1.16-2.01); P = 0.003).
Conclusion: Patients with early-onset PCa had distinct somatic gene tumor mutations in ERF and CDK12. Therapeutic targeting of genes associated with early-onset PCa can be potentially useful in future clinical studies.
Introduction
Early onset Prostate Cancer (PCa) patients’ incidence has increased sharply in the last two decades. The molecular biology of these tumors differs from the late onset presentation in terms of aggressiveness [1]. In this regard, the diagnosis of PCa at a young age is often an indication of more aggressive or fast-growing tumors. Patients diagnosed with early onset PCa have an increased risk of prostate cancer-specific mortality (PCSM) [2]. Most interestingly, the risk may be attributable to clinically significant tumors in patients diagnosed with high-risk and metastatic disease. Despite efforts to improve screening strategies, these patients are more likely to die from their cancers compared to those diagnosed later in life [3].
PCa can be further divided into early and late-onset, with men diagnosed at age 55 years or younger considered to be early-onset, making up over 10% of new diagnoses in 2012 [3]. Several theories have been proposed for the increasing incidence of early-onset PCa, essentially PSA screening in men under age 55, increased HPV prevalence, and exposure to environmental carcinogens. Additional factors involved include racial, ethnic, familial, and genetic factors [4].
In recent years, Next-Generation Sequencing (NGS) technologies have revolutionized cancer genomics research, providing a comprehensive view of genomic alterations associated with cancer development and progression [5]. Somatic testing may be performed to identify whether the tumor has actionable targets [6]. These findings can ultimately provide the clarity needed to best direct the clinical management for these patients and providers [7].
Despite the high incidence of PCa, the treatment options are limited. Therefore, NGS is gaining weight as a pharmacological target [8]. Several studies have reported its use to identify genetic alterations in prostate cancer, including somatic mutations in tumor suppressor genes such as TP53, PTEN and RB1, as well as alterations in androgen receptor signaling pathway genes such as AR and FOXA1 [9].
To our knowledge, no studies have compared molecular phenotypes of early versus late onset PCa. The article aims to use the international clinic-genomic data-sharing consortium from the Genomics Evidence Neoplasia Information Exchange (GENIE) to characterize somatic genetic profiles between patients with early and late-onset PCa and potential treatment.
Methods
Data source and study population
The American Association for Cancer Research Project GENIE registry contains sequencing data for more than 121,000 patients with cancer. Data is derived from 19 of the leading cancer centers around the world. GENIE importantly aggregates NGS data with patient demographics data and clinical outcomes. The study was exempted by our institutional review board because the data contained de-identified patient information. Patients were excluded with prostate small cell carcinoma (55 cases), prostate neuroendocrine carcinoma (53 cases), or prostate squamous cell carcinoma (2 cases). Patients with missing ages were removed (52 cases). A total of 4,546 Patients and 5,740 samples were included with pathologically confirmed prostate adenocarcinoma. The data is derived from 19 cancer centers from 2012 to 2021. This study adhered to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Variables
Patient demographic data included age at the time of diagnosis (in years), race (White, Black, Asian/Pacific Islander, Other, or Unknown), and Ethnicity (non-Spanish/non-Hispanic or Spanish/Hispanic). Patient clinical data included sample type (Primary Tumor, Metastasis, and Unknown) and the sequencing center.
Somatic gene mutation endpoints
The somatic gene mutations are derived from clinical-grade, NGS data from specimens obtained during routine patient care. Somatic mutations with a frequency among more than 2% of patients and with a minimum of 20 samples were included as a cutoff point for inclusion criteria. The primary endpoint in our study is the differential expression of somatic gene mutations.
Statistical analysis
Patients were dichotomized into early-onset (< 55 years old) and late-onset (> 55 years old) prostate cancer based on their age at the time of clinical sequencing. Differences between the two groups were assessed using the median and interquartile range for continuous variables while using frequency and percentage for categorical data. A multivariable logistic regression was performed to make comparisons of the variant gene mutations between early-onset and late-onset prostate cancer. The covariates in the logistic regression included: patient race, ethnicity, sequencing center, and primary sample type. Survival analysis was depicted using a weighted Kaplan-Meier plot and statistical significance was assessed by log rank test. Significance was defined for all tests using a two-tailed P - value of < 0.05. Analysis in this study was conducted using SPSS version 28.0 (IBM Corporation) and the RCommander package of R version 4.1.0.
Results
A total of 4092 adult male patients were recorded with PCa with data derived from 19 international cancer centers between 2012 and 2021 in the AACR Genie database. Overall, the patient population tended to be made up of non-Hispanic White (86.2%) men with primary tumors (63.6%). Patients had a mean age (SD) at clinical sequencing of 69.2 (7.7) years old. A summary of the patient demographics can be found in Table 1A. Patients were dichotomized into early-onset (< 55 years old) and late-onset (≥ 55 years old) PCa. A total of 452 patients (11.0%) were defined as having a diagnosis of early-onset PCa with a mean age (SD) of clinical sequencing at 51.4 (3.2) years old; a total of 3640 patients (89.0%) had a diagnosis of late-onset prostate cancer with a mean age (SD) of clinical sequencing of at 68.2 (7.1) years old. Patients with early-onset PCa were more likely to identify as Black (12.2% vs. 7.7%) but had lower rates of metastatic disease (32.0% vs. 45.0%) compared to late-onset prostate cancer in our population.
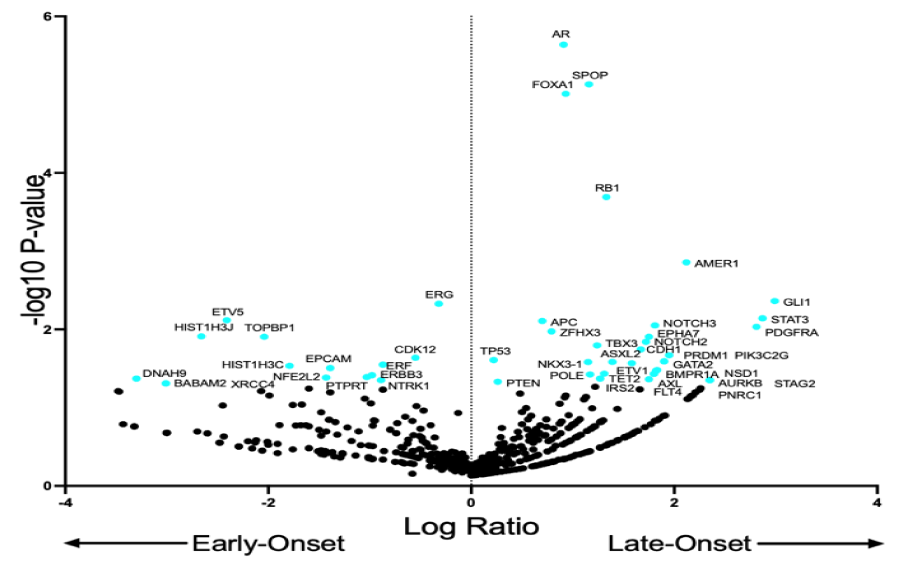
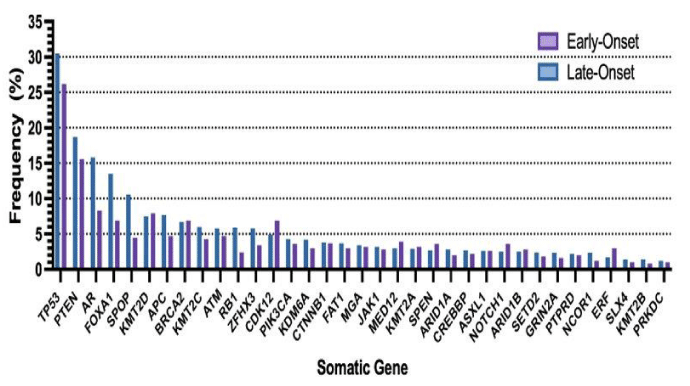
Among the 4552 samples examined, a total of 35 genes had a somatic gene mutation frequency greater than 1% of the profiled samples of prostate cancer specimens. The variations of the genomic samples included for both groups are shown in Figure 1. The frequency for each of the genes between early and late-onset PCa can be found in Figure 2. Approximately 3 out of every 10 patients had a TP53 mutation which is commonly recognized as a driver gene in prostate cancer. The genes PTEN, AR, and FOXA1 were all found in greater than 10% of the prostate cancer specimens while SPOP, KMT2D, APC, BRCA2, KMT2C, ATM, RB1, ZFHX3, and CDK12 were all found among greater than 5% of the prostate cancer specimens. A summary of the most significant (p < 0.001) co-occurring or mutually exclusive genes based on frequency is defined in Table 1B. Among both early and late-onset prostate cancer, the genes TP53 and CDK12 are mutually exclusive.
Baseline somatic gene variations are described for the entire population; somatic gene variations are further defined in early-onset and late-onset prostate cancer in Table 2.
A logistic regression was performed to control for differences in both patient and facility differences (race/ethnicity, sequencing center, and sample type). Patients with early-onset prostate cancer had higher odds of having a somatic mutation in CDK12 [1.51 (95% CI: 1.04-2.22)] or ERF [1.81 (95% CI: 1.02-3.24)] compared to patients who were diagnosed late in life. In contrast, patients with early-onset prostate cancer had lower odds of having a mutation in AR [0.64 (95% CI: 0.44-0.91); P = 0.014], FOXA1 [0.46 (95% CI: 0.32-0.66); P <0.001)], SPOP [0.37 (95% CI: 0.24-0.58); P <0.001], RB1 [0.48 (95% CI: 0.26-0.86); P = 0.014], and ZFHX3 [0.59 (95% CI: 0.36-0.99); P = 0.044].
A further subgroup analysis was performed to assess the genes associated with early-onset PCa. Patients with a CDK12 somatic gene variation were significantly more likely to be recorded by race as Black compared to White [1.92 (95% CI: 1.28- 2.86); P = 0.002). Moreover, patients with a CDK12 somatic gene variation were more likely to present with metastatic disease compared to primary prostate cancer [1.53 (95% CI: 1.16-2.01); P = 0.003). No significant differences were found among any of the patient demographics for patients with a somatic gene variation in ERF.
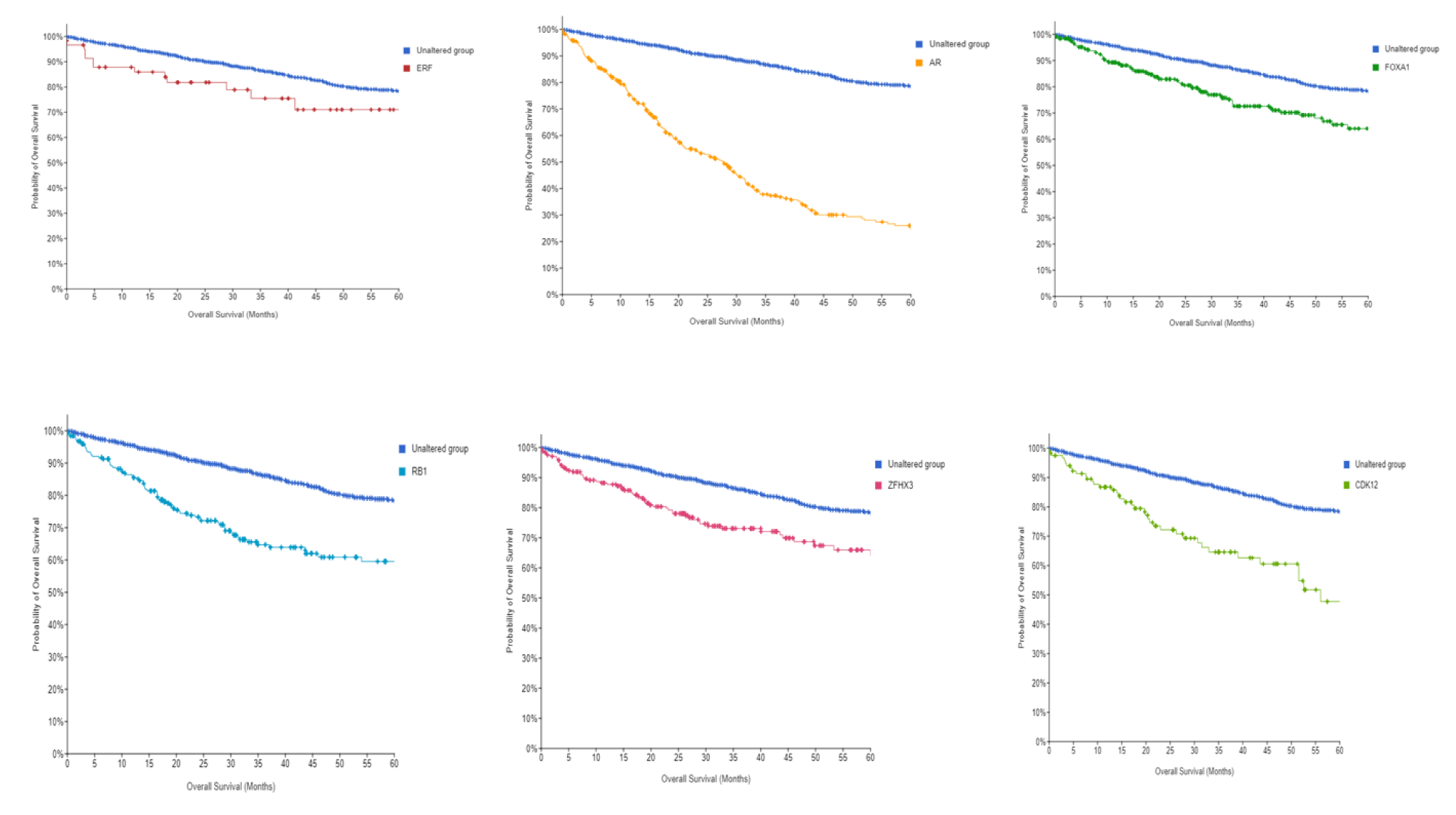
Interestingly, on survival analysis, the AR and CDK12 genes were associated with worse survival compared to the other genes in our panel. Figure 3 depicts the 5-year Kaplan-Meier curves for ERF, AR, FOXA1, RB1, ZFHX3, and CDK12. The median overall survival for the entire cohort was 141 months (95% CI 114-166, P < 0.001). The overall survival for AR and CDK12 genes was 30 months (95% CI 21-34, P < 0.001) and 56 months (95% CI 51-70, P < 0.001) respectively.
Discussion
This study used multivariate analysis to identify multiple genes associated with early and late-onset prostate cancer. Key genes associated with early-onset prostate cancer included CDK12 and ERF. Genes associated with this study for late-onset prostate cancer included AR, FOXA1, SPOP, and ZFHX3.
The genetic pathogenesis of prostate cancer
The genetic pathogenesis of PCa has been explored in multiple contexts, including germline, somatic, and epigenetic mutations. The collective knowledge base for all forms of genetic testing has rapidly increased in recent decades, as improved access to NGS tools and declining costs have allowed clinicians and researchers alike to benefit heavily from these resources [10,11]. A thorough family history (including history of previously identified high-risk germline mutations, certain ancestries, and multiple multi-organ cancers) has been the mainstay of initiating genetic assessment for clinicians and assessment of the genetic profiles of these patients has been essential in the recognition of pertinent germline mutations [12]. The identification of somatic mutations within active prostate cancers has also been a growing field of research and has functioned as a driver for optimizing and designing novel chemotherapies and immunotherapies [13]. Also included in this field of research has been the impact of multiple epigenetic drivers, such as chromatin remodeling, DNA methylation, and histone modification on the occurrence of prostate cancer and management of treatment resistance [14]. The absolute prevalence of prostate cancer and its increasing occurrence, especially among younger patients, underlies the importance of identifying the prognostic and therapeutic implications of prostate-cancer-related mutations. There exists a gap in the current literature in identifying the role of these markers differentially between early and late-onset PCa. The current study aims to clarify this gap by identifying the highest yield markers for early versus late onset prostate cancer.
CDK12
Cyclin-dependent kinase 12 (CDK12), encoded on chromosome 17q12, belongs to the cyclin-dependent kinase (CDK) family of serine/threonine protein kinases that regulate transcriptional and post-transcriptional processes, thereby having the ability to modulate multiple cellular functions [15]. CDK12 typically functions as a complex with cyclin K, with its most well-characterized roles in the regulation of gene transcription. Mutations in CDK12 have been associated with ovarian, breast, esophageal, uterine, bladder, stomach, colorectal, pancreatic, and prostate carcinomas [16]. CDK12 aberrations have been reported as a biomarker of response to immunotherapy for metastatic castration-resistant prostate cancer (mCRPC), with deleterious CDK12 alterations found in 2-4% of primary prostate cancers and 4.7% - 11% of mCRPC [17]. Common CDK12 mutations include tandem duplications, genomic rearrangements, missense alterations, nonsense alterations, and frameshift mutations or indels [18]. FDA-approved drugs used in patients with CDK12-mutated prostate cancer include abiraterone and enzalutamide, however, studies have also shown that patients with CDK12-altered prostate cancer respond to PD-1 inhibitors such as pembrolizumab [19]. An ongoing prospective clinical trial is currently examining the combination of ipilimumab plus nivolumab in patients with tumors possessing CDK12 alterations [20]. In our study, patients with early-onset PCa had higher odds of having a mutation in CDK12 compared to patients who were diagnosed later in life [1.51 (95% CI: 1.04-2.22); P = 0.032].
ERF
ERF, also known as ETS2 Repressor Factor, is a gene that functions to antagonize the members of the ETS protein family that collectively operate as transcription factors [21]. ERF has been identified to operate in tandem with ERG, an oncoprotein whose gene fusion product with TMPRSS2 is seen in nearly half of all prostate carcinomas [22]. The effects of a loss of function in ERF have been demonstrated to contribute to a tumor microenvironment similar to the effects seen by the gain of function in TMPRSS2–ERG [23]. Because of this, ERF and TMPRSS2–ERG work competitively, and an imbalance in either can promote a tumor-supportive environment [24]. Both deletions and mutations of ERF for PCa have been identified to occur frequently in African-American populations [25]. Quantification of ERF gene products may also provide value as a prognostic factor for PCa, as copy-number loss has been identified to be significantly associated with more aggressive pathologic features such as T staging, Gleason grading, and residual tumor burden in African American men [24,25]. While mutations in ERF are most often seen with prostate cancer, mutated ERF has also been found in other tumor types including colorectal adenocarcinoma and Ewing Sarcoma [26,27]. There are currently no therapies or active trials examining the use of ERF as a target for prostate cancer management. In this study, patients with early-onset prostate cancer had higher odds of having a mutation in ERF compared to patients who were diagnosed later in life [1.81 (95% CI: 1.02-3.24); P = 0.043]
AR
The majority of prostate cancers represent androgen-sensitive malignancies, characterized by their reliance on the transcriptional activity of the androgen receptor (AR) for growth and proliferation [28]. The AR signaling is the main pathway involved in PCa growth, making it a subject of intense scientific scrutiny and therapeutic focus [29]. Therapies designed to inhibit AR activity, such as androgen deprivation therapy and next-generation AR antagonists, have demonstrated significant clinical efficacy in managing this disease, underscoring the critical role of the androgen receptor in prostate cancer progression [30,31]. In the era of precision medicine, this involves detailed characterization of the AR pathway, including the presence of AR mutations, amplifications, and expression levels, as well as the potential presence of constitutively active AR variants. Such comprehensive molecular profiling enables oncologists to select the most suitable therapies, including AR-targeted agents, to precisely inhibit AR signaling and address the specific drivers of the disease in each patient [32,33]. In our analysis, AR mutations were more likely to be present in late-onset prostate cancer patients, out of which were likely to have worse overall survival compared to the patients without the mutation.
FOXA1, SPOP and ZFHX3
Emerging data links unfavorable outcomes to hotspot mutations in FOXA1 [34], while mutations in ZFHX3 and SPOP remain unclear [35]. FOXA1 is a transcription factor that modules the transcription of the AR and it is considered to have an important role in facilitating prostate cancer growth [36,37]. In our cohort, these mutations were more likely to be present in late-onset prostate cancer and in agreement with the current literature. The transcription factor known as zinc-finger homeobox 3 (ZFHX3), alternatively referred to as ATBF1 (AT motif-binding factor 1), is a sizable protein encompassing 23 zinc-finger domains, along with 4 homeodomains and various additional motifs [38]. ZFHX3 exhibits frequent mutations in cases of metastatic or high-grade human prostate cancers, with a notable portion of these mutations being frameshift alterations, ultimately leading to functional inactivation of the protein [39]. Inactivating point mutations within the gene responsible for encoding the speckle-type poxvirus and zinc-finger protein (SPOP) represent one of the prevailing genetic alterations in prostate cancer. These mutations are observed with notable frequency, occurring in approximately 6% to 15% of cases across both localized and metastatic forms of the disease [40]. The inactivation of SPOP leads to an outcome wherein there is an elevated expression of the Androgen Receptor (AR) at the protein level. This, in turn, augments AR-mediated cellular proliferation, emphasizing the pivotal role of SPOP in regulating AR-dependent pathways in prostate cancer [41]. Collectively, these discoveries have implicated SPOP alterations as a distinctive hallmark, delineating a novel subclass of prostate cancers [42].
Clinical implications
The analysis of mutations within our cohort has revealed a landscape characterized by several mutations across various genes based on age of onset. These mutations have the potential to profoundly impact the intricate signaling pathways that play pivotal roles in treatment, identifying such important mutations is the cornerstone to support clinical and treatment decisions. One notable example is the use of androgen receptor-targeted agents. In addition to AR-targeted therapies, other targeted agents are being investigated, such as PARP inhibitors for patients with DNA repair gene mutations and immunotherapies that aim to harness the immune system to attack cancer cells [43]. These approaches are tailored to the genetic and molecular characteristics of the patient’s cancer, representing a more personalized and effective treatment strategy. In our cohort. We identified that CDK12 and ERF were more likely to be mutated in early onset. In contrast to mutation in the AR receptor which was more frequently mutated in late-onset patients. Most interestingly, mutations in the AR, FOXA1, RB1, ZFHX3, and SPOP were more likely to have worse overall survival compared to patients without mutations. These findings are important to further our understanding of disease progression in prostate cancer.
Limitations
The source data used in this manuscript is directly from the AACR Project GENIE. The vast data sharing performed by this consortium allows this study to include a large number of verified, relevant data points. There are limitations, however, created by the use of GENIE that must be acknowledged. GENIE does not provide information about the stage, grade, or treatment history of tumors. Pertinent demographic factors such as lifestyle modifiers (i.e. obesity) or family history of cancer are also not available. Collectively, these factors represent possible contributors to age-related prostate cancer that this study was unable to explore. Additionally, the genes available for assessment within GENIE have established cancer markers genes that have previously been sequenced. This study does not identify or assess new, novel cancer genes. Further, there is an inherent bias in patient selection as not all institutions are part of AACR Project Genie and/or have access to the appropriate broad tumor sequencing infrastructure.
Conclusion
The study identified somatic mutations based on age of onset of prostate cancer. Mutations in CDK12 and ERF were more likely to be present in early onset. In contrast, genes associated with late-onset prostate cancer included AR, FOXA1, SPOP, and ZFHX3, which are considered to be actionable mutations with the evolution of gene-targeted therapy. These findings can aid with the current guidelines for age-related prostate cancer screening and management. The effective implementation of precision medicine in prostate cancer management necessitates the establishment of novel genetic biomarker classifications, allowing the stratification of patients into distinct subgroups tailored to receive specific therapies accustomed to their unique genetic profiles.
The authors would like to acknowledge the American Association for Cancer Research and its financial and material support in the development of the AACR Project GENIE registry, as well as members of the consortium for their commitment to data sharing. Interpretations are the responsibility of the study authors.
- Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: an important clinical entity. Nat Rev Urol. 2014 Jun;11(6):317-23. doi: 10.1038/nrurol.2014.91. Epub 2014 May 13. PMID: 24818853; PMCID: PMC4191828.
- Bleyer A, Spreafico F, Barr R. Prostate cancer in young men: An emerging young adult and older adolescent challenge. Cancer. 2020 Jan 1; 126(1):46-57.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209-249. doi: 10.3322/caac.21660. Epub 2021 Feb 4. PMID: 33538338.
- Bleyer A, Spreafico F, Barr R. Prostate cancer in young men: An emerging young adult and older adolescent challenge. Cancer. 2020 Jan 1;126(1):46-57. doi: 10.1002/cncr.32498. Epub 2019 Sep 25. PMID: 31553489.
- Cheng HH, Sokolova AO, Schaeffer EM, Small EJ, Higano CS. Germline and somatic mutations in prostate cancer for the clinician. Journal of the National Comprehensive Cancer Network. 2019 May 1;17(5):515-21.
- Ikeda S, Elkin SK, Tomson BN, Carter JL, Kurzrock R. Next-generation sequencing of prostate cancer: genomic and pathway alterations, potential actionability patterns, and relative rate of use of clinical-grade testing. Cancer Biol Ther. 2019;20(2):219-226. doi: 10.1080/15384047.2018.1523849. Epub 2018 Oct 19. PMID: 30339521; PMCID: PMC6343723.
- Saito M, Momma T, Kono K. Targeted therapy according to next generation sequencing-based panel sequencing. Fukushima J Med Sci. 2018 Apr 17;64(1):9-14. doi: 10.5387/fms.2018-02. Epub 2018 Apr 7. PMID: 29628467; PMCID: PMC5956085.
- Bose R, Karthaus WR, Armenia J, Abida W, Iaquinta PJ, Zhang Z, Wongvipat J, Wasmuth EV, Shah N, Sullivan PS, Doran MG, Wang P, Patruno A, Zhao Y; International SU2C/PCF Prostate Cancer Dream Team; Zheng D, Schultz N, Sawyers CL. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature. 2017 Jun 29;546(7660):671-675. doi: 10.1038/nature22820. Epub 2017 Jun 14. PMID: 28614298; PMCID: PMC5576182.
- Hovelson DH, Tomlins SA. The Role of Next-Generation Sequencing in Castration-Resistant Prostate Cancer Treatment. Cancer J. 2016 Sep/Oct;22(5):357-361. doi: 10.1097/PPO.0000000000000217. PMID: 27749331; PMCID: PMC5759046.
- Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour A, Rabasha B, Bahl S, Mullane SA, Robinson BD, Aldubayan S, Khani F, Karir B, Kim E, Chimene-Weiss J, Hofree M, Romanel A, Osborne JR, Kim JW, Azabdaftari G, Woloszynska-Read A, Sfanos K, De Marzo AM, Demichelis F, Gabriel S, Van Allen EM, Mesirov J, Tamayo P, Rubin MA, Powell IJ, Garraway LA. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer Discov. 2017 Sep;7(9):973-983. doi: 10.1158/2159-8290.CD-16-0960. Epub 2017 May 17. PMID: 28515055; PMCID: PMC5836784.
- Schwarze K, Buchanan J, Fermont JM, Dreau H, Tilley MW, Taylor JM, Antoniou P, Knight SJL, Camps C, Pentony MM, Kvikstad EM, Harris S, Popitsch N, Pagnamenta AT, Schuh A, Taylor JC, Wordsworth S. The complete costs of genome sequencing: a microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet Med. 2020 Jan;22(1):85-94. doi: 10.1038/s41436-019-0618-7. Epub 2019 Jul 30. PMID: 31358947; PMCID: PMC6944636.
- Pruneri G, De Braud F, Sapino A, Aglietta M, Vecchione A, Giusti R, Marchiò C, Scarpino S, Baggi A, Bonetti G, Franzini JM, Volpe M, Jommi C. Next-Generation Sequencing in Clinical Practice: Is It a Cost-Saving Alternative to a Single-Gene Testing Approach? Pharmacoecon Open. 2021 Jun;5(2):285-298. doi: 10.1007/s41669-020-00249-0. Epub 2021 Mar 4. PMID: 33660227; PMCID: PMC8160052.
- Doan DK, Schmidt KT, Chau CH, Figg WD. Germline Genetics of Prostate Cancer: Prevalence of Risk Variants and Clinical Implications for Disease Management. Cancers (Basel). 2021 Apr 29;13(9):2154. doi: 10.3390/cancers13092154. PMID: 33947030; PMCID: PMC8124444.
- Fay EK, Graff JN. Immunotherapy in Prostate Cancer. Cancers (Basel). 2020;12(7):1752. Published 2020 Jul 1. doi:10.3390/cancers12071752
- Goel S, Bhatia V, Biswas T, Ateeq B. Epigenetic reprogramming during prostate cancer progression: A perspective from development. Semin Cancer Biol. 2022 Aug;83:136-151. doi: 10.1016/j.semcancer.2021.01.009. Epub 2021 Feb 2. PMID: 33545340; PMCID: PMC7612861.
- Lui GYL, Grandori C, Kemp CJ. CDK12: an emerging therapeutic target for cancer. J Clin Pathol. 2018 Nov;71(11):957-962. doi: 10.1136/jclinpath-2018-205356. Epub 2018 Aug 13. PMID: 30104286; PMCID: PMC6242340.
- Paculová H, Kohoutek J. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017 Oct 27;12:7. doi: 10.1186/s13008-017-0033-x. PMID: 29090014; PMCID: PMC5658942.
- Rescigno P, Gurel B, Pereira R, Crespo M, Rekowski J, Rediti M, Barrero M, Mateo J, Bianchini D, Messina C, Fenor de la Maza MD, Chandran K, Carmichael J, Guo C, Paschalis A, Sharp A, Seed G, Figueiredo I, Lambros M, Miranda S, Ferreira A, Bertan C, Riisnaes R, Porta N, Yuan W, Carreira S, de Bono JS. Characterizing CDK12-Mutated Prostate Cancers. Clin Cancer Res. 2021 Jan 15;27(2):566-574. doi: 10.1158/1078-0432.CCR-20-2371. Epub 2020 Sep 28. PMID: 32988971; PMCID: PMC7855716.
- Pan E, Cabal A, Javier-DesLoges J, Patel D, Panian J, Lee S, Shaya J, Nonato T, Xu X, Stewart T, Rose B, Shabaik A, Cohen E, Kurzrock R, Tamayo P, McKay RR. Analysis of CDK12 alterations in a pan-cancer database. Cancer Med. 2022 Feb;11(3):753-763. doi: 10.1002/cam4.4483. Epub 2021 Dec 12. PMID: 34898046; PMCID: PMC8817093.
- Antonarakis ES, Isaacsson Velho P, Fu W, Wang H, Agarwal N, Sacristan Santos V, Maughan BL, Pili R, Adra N, Sternberg CN, Vlachostergios PJ, Tagawa ST, Bryce AH, McNatty AL, Reichert ZR, Dreicer R, Sartor O, Lotan TL, Hussain M. CDK12-Altered Prostate Cancer: Clinical Features and Therapeutic Outcomes to Standard Systemic Therapies, Poly (ADP-Ribose) Polymerase Inhibitors, and PD-1 Inhibitors. JCO Precis Oncol. 2020;4:370-381. doi: 10.1200/po.19.00399. Epub 2020 Apr 21. PMID: 32462107; PMCID: PMC7252221.
- Schweizer MT, Ha G, Gulati R, Brown LC, McKay RR, Dorff T, Hoge ACH, Reichel J, Vats P, Kilari D, Patel V, Oh WK, Chinnaiyan A, Pritchard CC, Armstrong AJ, Montgomery RB, Alva A. CDK12-Mutated Prostate Cancer: Clinical Outcomes With Standard Therapies and Immune Checkpoint Blockade. JCO Precis Oncol. 2020;4:382-392. doi: 10.1200/po.19.00383. Epub 2020 Apr 21. PMID: 32671317; PMCID: PMC7363399.
- Sgouras DN, Athanasiou MA, Beal GJ Jr, Fisher RJ, Blair DG, Mavrothalassitis GJ. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995 Oct 2;14(19):4781-93. doi: 10.1002/j.1460-2075.1995.tb00160.x. PMID: 7588608; PMCID: PMC394576.
- Bose R, Karthaus WR, Armenia J, Abida W, Iaquinta PJ, Zhang Z, Wongvipat J, Wasmuth EV, Shah N, Sullivan PS, Doran MG, Wang P, Patruno A, Zhao Y; International SU2C/PCF Prostate Cancer Dream Team; Zheng D, Schultz N, Sawyers CL. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature. 2017 Jun 29;546(7660):671-675. doi: 10.1038/nature22820. Epub 2017 Jun 14. PMID: 28614298; PMCID: PMC5576182.
- Lorenzin F, Demichelis F. Past, Current, and Future Strategies to Target ERG Fusion-Positive Prostate Cancer. Cancers (Basel). 2022 Feb 22;14(5):1118. doi: 10.3390/cancers14051118. PMID: 35267426; PMCID: PMC8909394.
- Zhou F, Gao S, Han D, Han W, Chen S, Patalano S, Macoska JA, He HH, Cai C. TMPRSS2-ERG activates NO-cGMP signaling in prostate cancer cells. Oncogene. 2019 May;38(22):4397-4411. doi: 10.1038/s41388-019-0730-9. Epub 2019 Feb 4. PMID: 30718921; PMCID: PMC6542710.
- Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour A, Rabasha B, Bahl S, Mullane SA, Robinson BD, Aldubayan S, Khani F, Karir B, Kim E, Chimene-Weiss J, Hofree M, Romanel A, Osborne JR, Kim JW, Azabdaftari G, Woloszynska-Read A, Sfanos K, De Marzo AM, Demichelis F, Gabriel S, Van Allen EM, Mesirov J, Tamayo P, Rubin MA, Powell IJ, Garraway LA. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer Discov. 2017 Sep;7(9):973-983. doi: 10.1158/2159-8290.CD-16-0960. Epub 2017 May 17. PMID: 28515055; PMCID: PMC5836784.
- Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016 Apr 26;15(4):857-865. doi: 10.1016/j.celrep.2016.03.075. Epub 2016 Apr 14. Erratum in: Cell Rep. 2016 Oct 18;17 (4):1206. PMID: 27149842; PMCID: PMC4850357.
- Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS, Thorner AR, de Torres C, Lavarino C, Suñol M, McKenna A, Sivachenko A, Cibulskis K, Lawrence MS, Stojanov P, Rosenberg M, Ambrogio L, Auclair D, Seepo S, Blumenstiel B, DeFelice M, Imaz-Rosshandler I, Schwarz-Cruz Y Celis A, Rivera MN, Rodriguez-Galindo C, Fleming MD, Golub TR, Getz G, Mora J, Stegmaier K. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014 Nov;4(11):1326-41. doi: 10.1158/2159-8290.CD-13-1037. Epub 2014 Sep 3. PMID: 25186949.
- Fujita K, Nonomura N. Role of Androgen Receptor in Prostate Cancer: A Review. World J Mens Health. 2019 Sep;37(3):288-295. doi: 10.5534/wjmh.180040. Epub 2018 Sep 10. PMID: 30209899; PMCID: PMC6704300.
- Culig Z, Santer FR. Androgen receptor signaling in prostate cancer. Cancer Metastasis Rev. 2014 Sep;33(2-3):413-27. doi: 10.1007/s10555-013-9474-0. PMID: 24384911.
- Aurilio G, Cimadamore A, Mazzucchelli R, Lopez-Beltran A, Verri E, Scarpelli M, Massari F, Cheng L, Santoni M, Montironi R. Androgen Receptor Signaling Pathway in Prostate Cancer: From Genetics to Clinical Applications. Cells. 2020 Dec 10;9(12):2653. doi: 10.3390/cells9122653. PMID: 33321757; PMCID: PMC7763510.
- Sharifi N, Gulley JL, Dahut WL. Androgen Deprivation Therapy for Prostate Cancer. JAMA. 2005; 294(2):238-244. doi:10.1001/jama.294.2.238
- Morova T, McNeill DR, Lallous N, Gönen M, Dalal K, Wilson DM 3rd, Gürsoy A, Keskin Ö, Lack NA. Androgen receptor-binding sites are highly mutated in prostate cancer. Nat Commun. 2020 Feb 11;11(1):832. doi: 10.1038/s41467-020-14644-y. PMID: 32047165; PMCID: PMC7012874.
- Shiota M, Akamatsu S, Tsukahara S, Nagakawa S, Matsumoto T, Eto M. Androgen receptor mutations for precision medicine in prostate cancer. Endocr Relat Cancer. 2022 Aug 17;29(10):R143-R155. doi: 10.1530/ERC-22-0140. PMID: 35900853.
- Zhao Y, Tindall DJ, Huang H. Modulation of androgen receptor by FOXA1 and FOXO1 factors in prostate cancer. Int J Biol Sci. 2014 Jun 5;10(6):614-9. doi: 10.7150/ijbs.8389. PMID: 24948874; PMCID: PMC4062954.
- Mangolini A, Rocca C, Bassi C, Ippolito C, Negrini M, Dell'Atti L, Lanza G, Gafà R, Bianchi N, Pinton P, Aguiari G. Detection of disease-causing mutations in prostate cancer by NGS sequencing. Cell Biol Int. 2022 Jul;46(7):1047-1061. doi: 10.1002/cbin.11803. Epub 2022 Apr 6. PMID: 35347810; PMCID: PMC9320837.
- Gao S, Chen S, Han D, Barrett D, Han W, Ahmed M, Patalano S, Macoska JA, He HH, Cai C. Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Res. 2019 Sep;29(9):770-772. doi: 10.1038/s41422-019-0203-2. Epub 2019 Jul 19. PMID: 31324883; PMCID: PMC6796877.
- Shah N, Brown M. The sly oncogene: FOXA1 mutations in prostate cancer. Cancer Cell. 2019; 36: 119-121. https://doi.org/10.1016/j.ccell.2019.07.005
- Miura Y, Tam T, Ido A, Morinaga T, Miki T, Hashimoto T, Tamaoki T. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J Biol Chem. 1995 Nov 10;270(45):26840-8. doi: 10.1074/jbc.270.45.26840. PMID: 7592926.
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012 Jul 12;487(7406):239-43. doi: 10.1038/nature11125. PMID: 22722839; PMCID: PMC3396711.
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012 May 20;44(6):685-9. doi: 10.1038/ng.2279. PMID: 22610119; PMCID: PMC3673022.
- Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, Fiskus W, Rajendran M, Chew SA, Zimmermann M, Bond R, He B, Coarfa C, Mitsiades N. Correction: Androgen Receptor Is the Key Transcriptional Mediator of the Tumor Suppressor SPOP in Prostate Cancer. Cancer Res. 2019 Sep 1;79(17):4552. doi: 10.1158/0008-5472.CAN-19-1981. Erratum for: Cancer Res. 2014 Oct 1;74(19):5631-43. PMID: 31481421.
- Blattner M, Lee DJ, O'Reilly C, Park K, MacDonald TY, Khani F, Turner KR, Chiu YL, Wild PJ, Dolgalev I, Heguy A, Sboner A, Ramazangolu S, Hieronymus H, Sawyers C, Tewari AK, Moch H, Yoon GS, Known YC, Andrén O, Fall K, Demichelis F, Mosquera JM, Robinson BD, Barbieri CE, Rubin MA. SPOP mutations in prostate cancer across demographically diverse patient cohorts. Neoplasia. 2014 Jan;16(1):14-20. doi: 10.1593/neo.131704. PMID: 24563616; PMCID: PMC3924544.
- Congregado B, Rivero I, Osmán I, Sáez C, Medina López R. PARP Inhibitors: A New Horizon for Patients with Prostate Cancer. Biomedicines. 2022 Jun 15;10(6):1416. doi: 10.3390/biomedicines10061416. PMID: 35740437; PMCID: PMC9220343.