Journal of BioData Mining
Identification of Key Gene Modules and Potential ceRNA Network in Progressive Coronary Artery Disease by Weighted Gene Co-Expression Network Analysis
Zhiwei Gao1#, Chao Yan1#, Xiangkun Meng1#, Zhichao Jiang2#, Sen Li1, Gang Hu3, Haolong Shi4, Xinchen Huang5* and Jinren Zhou1*
1Department of Vascular Surgery, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, PR China
2Department of General Surgery, The Second People’s Hospital of Linhai, China
3Department of General Surgery, Zhejiang Yuyao People’s Hospital, China
4Center of Basic and Translational Research, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, PR China
5Department of Radiology, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China
#These authors have contributed equally to this work and share the first authorship
Jinren Zhou, Department of Vascular Surgery, The Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, PR China, E-mail: [email protected]
Cite this as
Gao Z, Yan C, Meng X, Jiang Z, Li S, Hu G, et al. Identification of Key Gene Modules and Potential ceRNA Network in Progressive Coronary Artery Disease by Weighted Gene Co-Expression Network Analysis. J BioData Min. 2025;1(1): 001-011. Available from: 10.17352/jbdm.000001Copyright Licence
© 2025 Gao Z, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Coronary Artery Disease (CAD) is a global chronic inflammatory disease with high morbidity and mortality, seriously endangering human health and life quality. Therefore, exploring the critical molecular mechanisms and identifying potential signaling pathways in CAD progression is vital. We reanalyzed peripheral blood mRNA microarray expression data from the GSE34822 dataset and identified 15 gene co-expression modules using weighted gene co-expression network analysis (WGCNA). One of the modules was found to be closely associated with CAD progression, mediating pathways such as platelet degranulation, platelet activation, and platelet aggregation. Genes including NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3 were significantly correlated with CAD progression. Positive regulation of interleukin−13 production and regulation of monocyte differentiation were identified to be related to these six key genes. Specifically, we discovered that the SMIM3 gene was associated with monocyte infiltration and further developed a SMIM3-related competitive endogenous RNA (ceRNA) network. This suggests that SMIM3 plays a role in monocyte differentiation, contributing to plaque instability and accelerating CAD progression. In this study, we identified six key genes in the crucial module as potential biomarkers for diagnosing and treating progressive CAD. Additionally, we constructed a ceRNA network offering insights into CAD’s underlying regulatory mechanisms.
Highlights
- NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3 provide significant association with CAD progression.
- SMIM3 was upregulated in patients with progressive CAD by qRT-PCR.
- Positive regulation of interleukin−13 production and regulation of monocyte differentiation were related to SMIM3 expression.
- The SMIM3 gene was associated with monocyte cell infiltration in patients with CAD progression.
Introduction
Coronary artery disease (CAD) is an inflammatory atherosclerotic disease that has become a significant global public health burden, manifesting as stable angina, unstable angina, myocardial infarction, or sudden cardiac death [1]. The formation of atherosclerotic plaques involves inflammation, leading to plaque erosion or rupture, which can cause partial or total coronary artery occlusion. Plaque buildup ultimately results in acute myocardial infarction [2,3]. The risk factors of CAD mainly include hypertension, diabetes, a sedentary lifestyle, smoking, obesity, stress, and hyperlipidemia [4]. Due to poor living and working habits, CAD still maintains high morbidity and mortality [5]. Understanding the mechanisms of plaque progression indicates that atherosclerosis is a complex and multifactorial disease [6]. CAD begins with damage to the lining of the coronary arteries, followed by the accumulation of fatty deposits composed of cholesterol and other cellular impurities at the injury site [7]. Specifically, low-density lipoprotein (LDL) plays a key role in the early events of atherosclerotic plaque formation. Accumulated LDL undergoes oxidative modification under the action of smooth muscle cells (SMCs), monocytes, and endothelial cells, forming oxidized LDL (ox-LDL). Ox-LDL further aggravates endothelial injury and stimulates endothelial cells to produce molecules such as E-selectin or P-selectin, promoting the migration of circulating monocytes to the vascular wall and promoting inflammation [8]. If the plaque surface ruptures, platelets accumulate at the injury site and try to repair the artery [9]. Clinically, CAD is considered a chronic disease; however, it is progressive, and clinical symptoms may not be obvious [1]. Stable CAD can remain stable for extended periods but can become unstable at any time due to plaque rupture or erosion, leading to acute coronary artery events [10]. In recent years, the prevention and treatment of CAD have remained a significant challenge worldwide. Therefore, an accurate assessment of stable CAD is urgent, and the molecular mechanisms associated with CAD progression must be explored further.
Weighted Gene Co-expression Network Analysis (WGCNA) is a mainstream research method primarily applied to large-sample gene expression data [11]. Genes are gathered together and grouped into a few modules based on gene expression similarity, and the modules are distinguished by the module eigengene or hub gene. Then the correlation between modules and clinical traits is calculated to select modules that are highly positively correlated to characteristics, and analyze the genes in the modules. Due to its excellent function, WGCNA has also been used in cardiovascular diseases [12,13], successfully identifying several potential genes or disease-related pathways. However, detecting the molecular mechanisms and signaling pathways of progressive CAD has not been reported.
To further reveal the potential key genes and signaling pathways in CAD progression, we constructed a weighted gene co-expression network using the GSE34822 dataset. We identified representative modules closely related to CAD progress. Vital regulatory genes were obtained by intersecting differentially expressed genes and the genes in usual modules. Then, functional enrichment analysis was used to identify the role of the interested genes in the pathological process, and gene set enrichment analysis (GSEA) [14] was used to reveal the potential regulatory processes of key genes. The CIBERSORT method was employed to assess the differential immune cell infiltration between progressive CAD samples and stable donors [15]. Finally, based on the predicted interactions of microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), we constructed competitive endogenous RNA (ceRNA) networks to further explore the mechanisms driving disease progression. This study enhances our understanding of CAD pathogenesis and offers valuable insights for potential therapeutic strategies.
Materials and methods
Data collection and analysis
The CAD-related dataset was obtained from GSE34822 [16]. The dataset comprised a total of 32 samples: 16 patients with significant lesion progression leading to repeated coronary interventions and 16 patients with angiographically documented stable courses. After computing and ranking the median absolute deviation, we filtered the top 75% of rank genes for WGCNA analysis. To identify differentially expressed genes (DEGs) in the GSE34822 microarray dataset, we employed the limma package (version 3.46.0) [17] in the R software. The selection criteria for DEGs were set to an adjusted p-value (adj.p.Val) < 0.05 and an absolute log2 fold change (|log2FC|) > 0.5. Furthermore, we obtained additional mRNA expression profiles of peripheral blood monocytes at different stages of CAD from GSE166780 [18]. The dataset consisted of 8 samples representing normal coronary arteries, 8 with intermediate coronary lesions, and 8 with acute myocardial infarction.
Weighted gene co-expression network analysis
The WGCNA R package (version 1.70.3) [11] was used to construct the network of 14,692 genes obtained from the GSE34822 dataset. To ensure reliable network construction results, we removed the outlier samples based on the unsupervised clustering method. The soft threshold power was selected based on the standard scale-free network criteria, and genes in the third quartile of variance were computed using a power function. MergeCutHeight referred to the height for cutting the dendrogram in the process of module merging, which was associated with the quantities and accuracy of modules we finally collected. Setting abline to 0.25, an appropriate quantity of modules was harvested. Eventually, the modules containing highly homogenous genes were merged under a 0.25 MergeCutHeight, and 15 modules were finally gathered. The grey module contained meaningless genes that cannot be categorized in any module by default and were subsequently removed in downstream analysis.
Identification of key gene modules associated with progressive CAD
Pearson’s correlation tests were conducted to evaluate the relationships between clinical traits and gene modules, helping to identify significant modules. We then defined Module Membership (MM) as the correlation between gene expression profiles and module eigengenes, while Gene Significance (GS) was defined as the absolute correlation between external traits and gene expression profiles. Further analyses were focused on genes within the most relevant modules, specifically those with the highest MM and GS values.
Functional enrichment analysis performed on interested modules
To identify the biological role that genes played in the pathological process, the clusterProfiler R package (version 4.0.0) [19] was utilized to carry out GO and KEGG enrichment analysis on targets we extracted from interested modules.
Gene set enrichment analysis
To investigate the underlying functions of the filtering critical genes involved in CAD with progression, we used the clusterProfiler R package (version 4.0.0) [19] for functional enrichment analysis based on GO biology process gene sets. The p value at < 0.05 was regarded as statistically enriched.
Immune infiltration analysis via CIBERSORT
To explore the composition of individual immune cells in CAD samples, we used the CIBERSORT tool for immune infiltration estimation [15]. CIBERSORT is a deconvolution method that characterizes the composition of the immune cells from gene expression profiles.
Receiver operating characteristic curve analysis
To validate the abilities of interested genes or factors for diagnosis, we used the pROC R package (version 1.17.0.1) [20] for constructing the Receiver Operating Characteristic (ROC) curve. The values of areas under the ROC curves were defined as AUC.
Construction of ceRNA Networks
We used two independent miRNA databases, respectively, Targetscan (https://www.targetscan.org/vert_80/) [21] and miRDB (http://mirdb.org/) [22], to predict target miRNAs of interested genes and screened common miRNAs in these two databases. Then, we used online StarBase database (https://starbase.sysu.edu.cn/index.php) [23] to predict lncRNAs that interacted with the predicted miRNAs based on the following screening criteria, including mammalian, human hg19 genome, strict stringency (≥ 5) of CLIP- Data, and with or without degradome data. CeRNA networks were constructed based on predicted interactions among mRNAs, miRNAs, and lncRNAs, and visualized using the Cytoscape software (version 3.8.2) [24].
Sample collection
Patients with progressive (n = 6) or stable (n = 6) CADs were selected from the second affiliated hospital of Zhejiang University School of Medicine, depending on whether there was evidence of disease progression. PERIPHERAL BLOOD MONONUCLEAR CELLS (PBMCs) were extracted from the heparinized venous blood of patients by Ficoll-Hypaque (Amersham Biosciences) density gradient centrifugation. All patients agreed to blood preservation after providing informed consent. This study involves human participants and was approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang University (I20211258 (1)).
Quantitative real-time PCR
RNA was extracted from patient PBMCs using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. cDNA was synthesized using reverse transcription kit (Roche, Indianapolis, IN, USA) and subsequently amplified by qRT-PCR using an ABI Prism system (Applied Biosystems, Foster City, CA). Data were analyzed using the ΔCt method normalized to GAPDH expression values in the samples. The measurements of each sample were performed in triplicate. The primer sequences are as follows, which were designed according to NCBI database and used for PCR amplification: NID2, 5′-CCGGTGCTGTCGTCGTTAC-3′ and 5′- GGCTTCGTAGAAGTGCAGGG-3′; PGRMC1, 5′-AAAGGCCGCAAATTCTACGG-3′ and 5′-CCCAGTCACTCAGAGTCTCCT-3′; TSC22D1, 5′-TCTCCGCTAGTATCAGCTCTAAC-3′ and 5′-ACACATCAAGGATCTCCGAAGAA-3′; LOC340508, 5′-GCAAGGATGGAGATGGGC-3′ and 5′-CTGGGTTTGGACTGGGAA-3′; KIAA1211, 5′-GAGGATCTGTTCCTGACCAGT-3′ and 5′-GGACTTAGAGAACTTGGCGTATC-3′; SMIM3, 5′-ATGGATGCAGTCAGCCAAGTC-3′ and 5′-CCCACTAAGGATCGGATGAGT-3′. All experiments were performed in accordance with relevant guidelines and regulations.
Statistics
All statistical analyses in this study were calculated using R software (http://www.r-project.org, version 4.0.4). The Wilcoxon rank-sum test was used to compare differences between two groups, while the Kruskal-Wallis H test was employed for comparisons among three groups. It was regarded as statistically significant when p was at < 0.05.
Results
Construction of weighted gene co-expression network in CAD
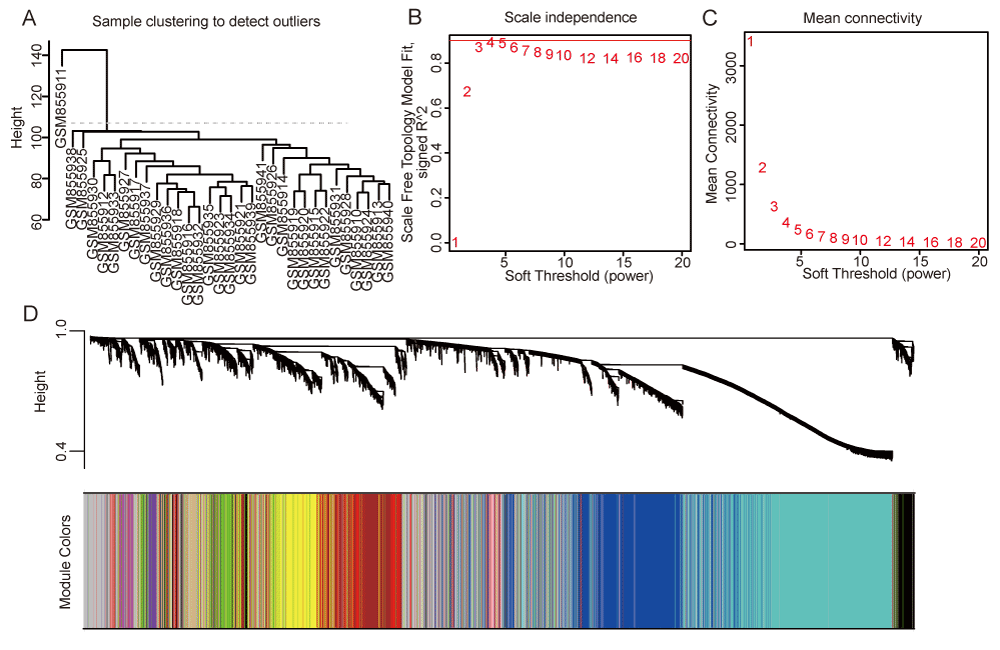
We collected microarray sequencing data from the peripheral blood of 32 patients with progressive and stable CAD, including 16 patients with progressive and 16 with stable disease in GSE34822. Sample GSM855911 was identified as an outlier and excluded from subsequent analyses (Figure 1A). To construct a scale-free topology network, the soft threshold power (β) of 3 was estimated (Figure 1B,1C). The hierarchical clustering tree revealed that 15 co-expression modules were identified (Figure 1D, S1A, TableS2).
Identification of modules in the co-expression network
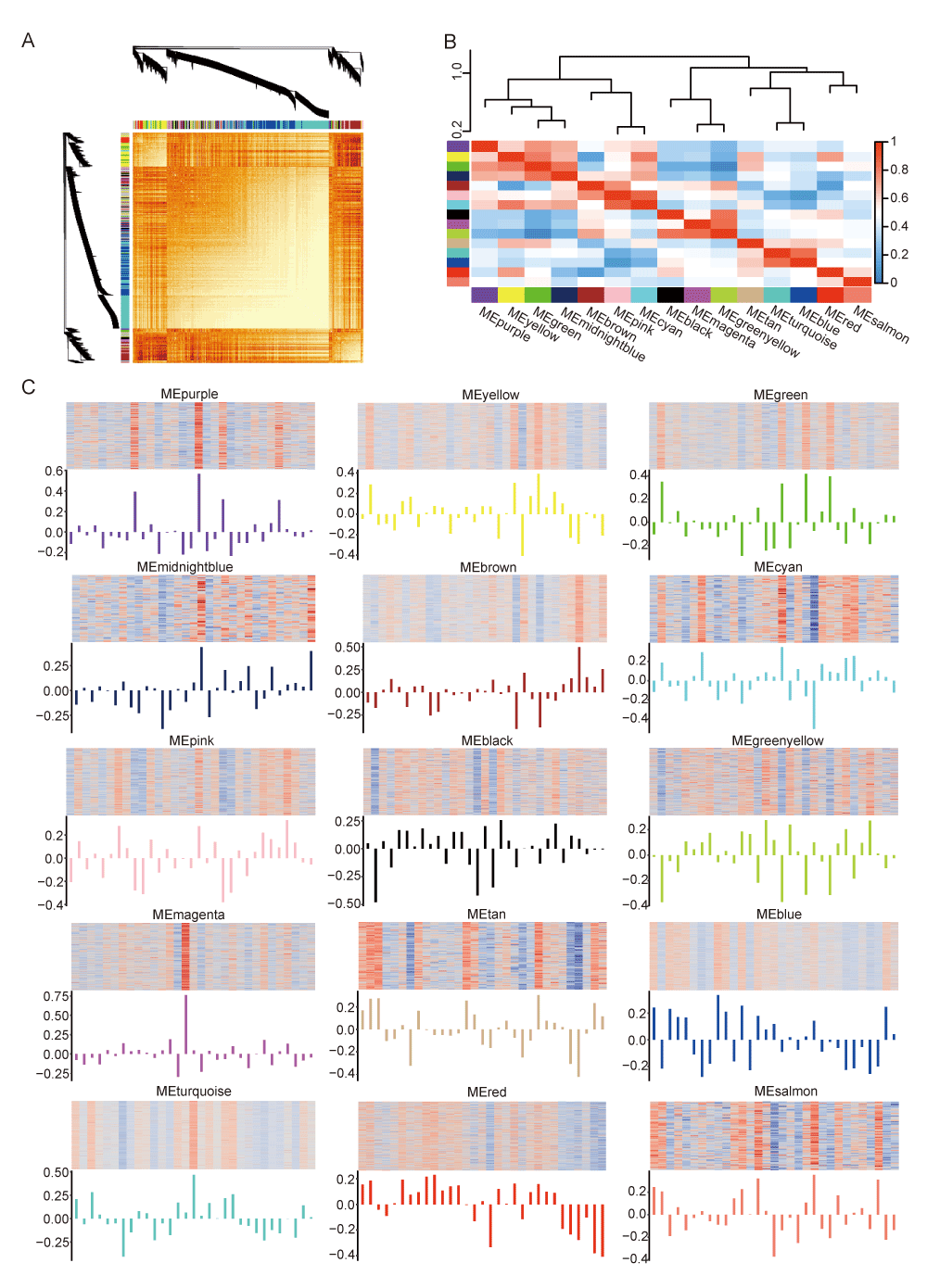
Following the construction of the weighted gene co-expression network, we calculated the dissTOM matrix (Figure 2A). Modules of correlated eigengenes were identified through a hierarchical clustering dendrogram and heatmap construction (Figure 2B). Figure 2C illustrates the expression of related genes and eigengene across the fifteen modules, excluding the grey module, which contained genes that could not be clustered.
Identification of gene co-expression modules correlated with progressive CAD traits
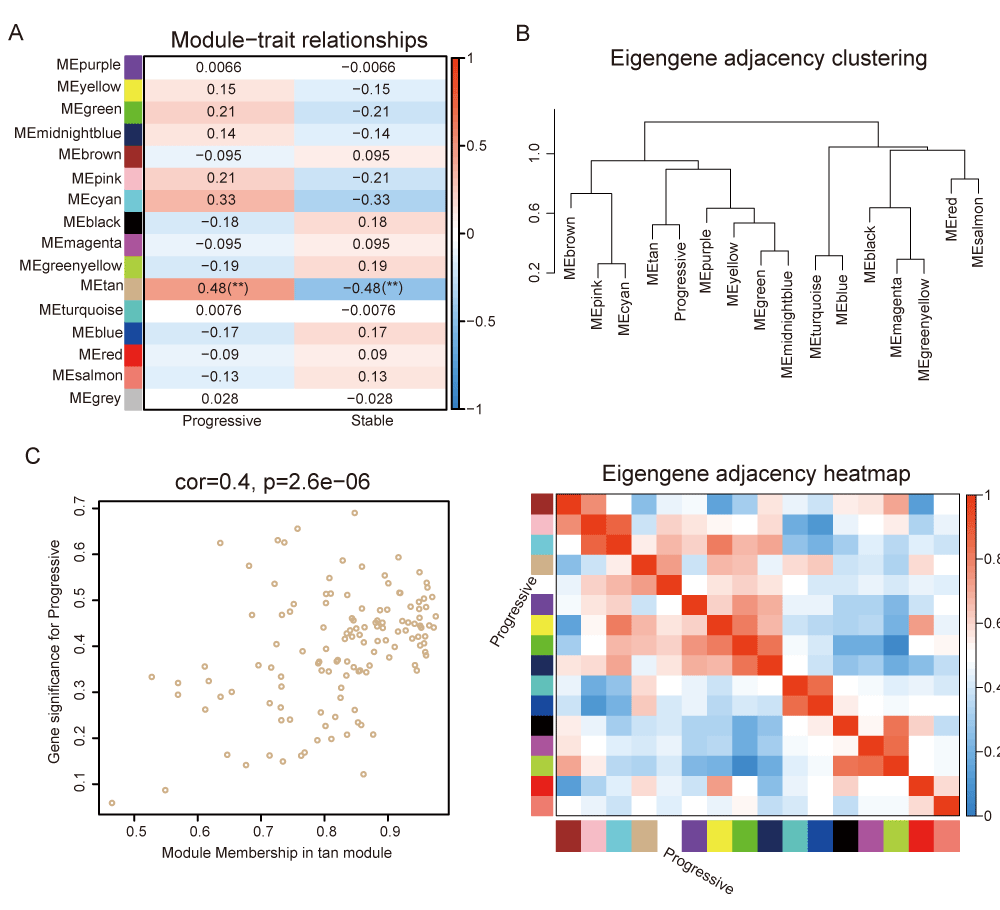
The Pearson correlation coefficients for the above modules and the clinical information were calculated to identify which modules were related to the clinical traits (Figure 3A and Table S3). The tan module showed a significant association with CAD progression (R = 0.48, p = 0.007) (Figure 3A), suggesting its potential biological significance in promoting CAD progression. Hierarchical clustering and heatmap analysis further confirmed that the tan module was most significantly associated with disease progression (Figure 3B). In addition, Figure 3C showed the significance of these genes in the tan module for disease progression (Rho = 0.4, p < 0.001).
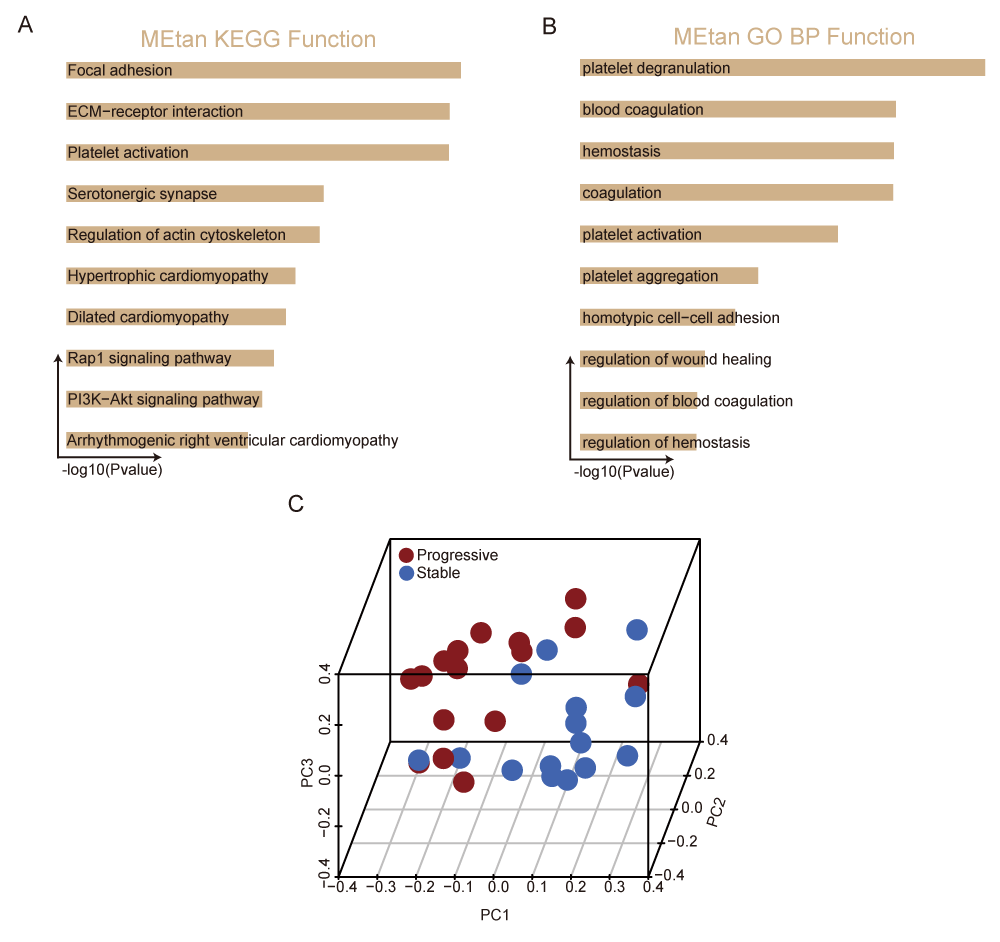
To further investigate the biological functions of the tan module, GO and KEGG pathway analyses were performed. Genes in the tan module were enriched in KEGG pathways involved in platelet degranulation, platelet activation, and platelet aggregation. In terms of biological process-associated pathways, these genes were mainly enriched in focal adhesion, ECM−receptor interaction, and platelet activation (Figure 4A,B, and Table S4). This suggested that platelet activation and aggregation play a crucial role in the progression of CAD [25]. To further verify the ability to diagnose progressive CAD, we could distinguish progressive CAD patients from stable CAD patients by principal component analysis based on the tan module eigengenes expression (Figure 4C). Furthermore, the ROC curve verified that the tan module could be employed to distinguish progressive CAD patients from stable CAD patients (AUC = 0.8042) (Figure S1B). Consequently, hub genes were identified within the tan module.
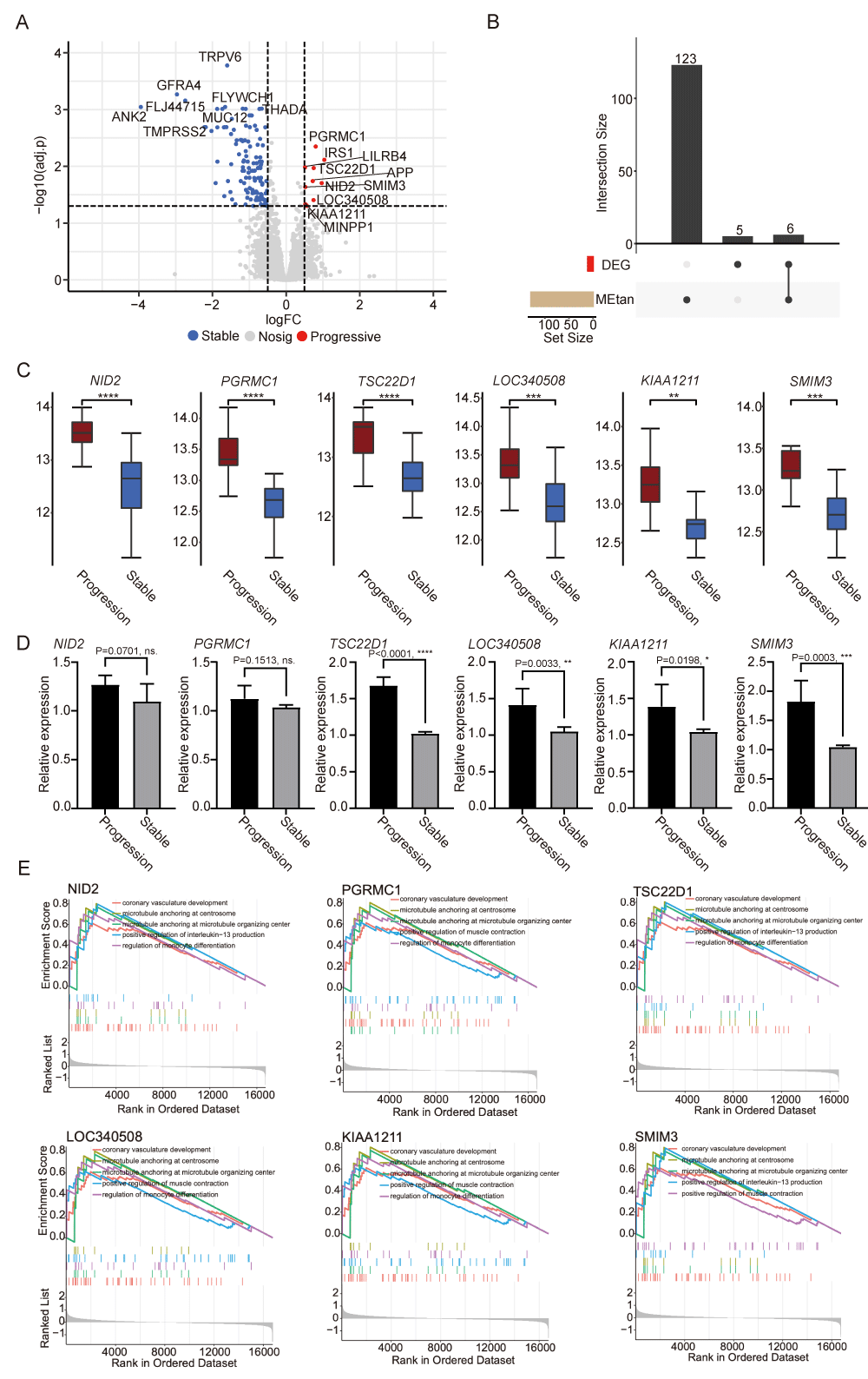
Authentication of key genes of tan module
To identify hub genes within the tan module, we compared progressive samples with stable samples and identified a total of 131 differentially expressed genes, which comprised 120 downregulated genes and 11 upregulated genes by analyzing the dataset GSE34822 (Figure 5A). Intersecting with the genes in the tan module, we obtained six key genes, including NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3 (Figure 5B). The expression of the six key genes was significantly and specifically upregulated in progressive samples (Figure 5C). Additionally, we quantified the ROC curves for each key gene to assess their diagnostic potential. The AUC values were as follows: PGRMC1 (0.9083), TSC22D1 (0.9042), NID2 (0.8875), LOC340508 (0.8583), KIAA1211 (0.8292), and SMIM3 (0.8667) (Figure S1C). These high AUC values indicate strong diagnostic potential. PBMC samples were collected from three patients with progressive CAD and three patients with stable CAD, and qPCR experiments were performed to verify the mRNA expression levels of the above six key genes (Figure 5D).
Functional enrichment analysis of CAD progressive key genes
To explore the specific functions of the identified key genes, we conducted GSEA analysis. As indicated by the results of the GSEA analysis, coronary vasculature development, microtubule anchoring at the centrosome, microtubule anchoring at the microtubule organizing center, positive regulation of interleukin−13 production, and regulation of monocyte differentiation were associated with these six key genes (Figures 5E and Table S5). These pathways had a close correlation with CAD, suggesting that the six key genes may play a role in CAD progression.
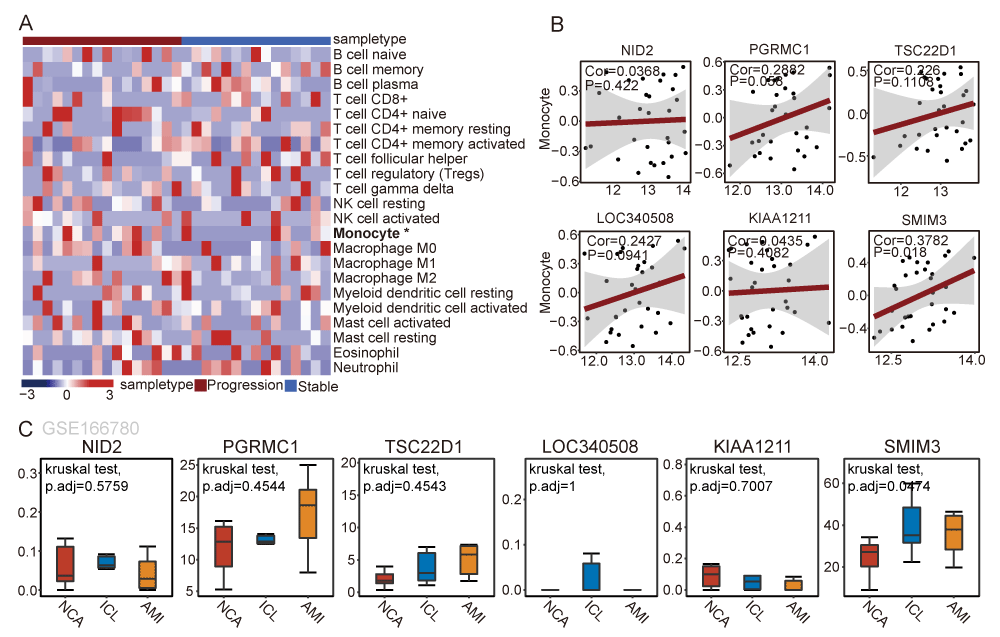
Assessment of immunocytes infiltration in CAD samples
The above findings suggest a close relationship between monocytes and CAD progression. We used the CIBERSORT algorithm to identify immune cell infiltration of patients suffering from progressive samples compared to stable samples. The results showed that monocytes were specifically enriched in the peripheral blood of patients with CAD progression (Figure 6A). We observed a significant positive correlation between the expression of SMIM3 and the composing proportion of Monocyte via correlation analysis (Rho = 0.3782, p = 0.018) (Figure 6B). The result suggested that patients with high expression of SMIM3, along with the enrichment of monocytes in peripheral blood, contribute to plaque instability and promote rapid CAD progression. We further verified the expression of 6 critical genes in peripheral blood monocytes in different stages of CAD. Further verification using the GSE166780 dataset showed consistent upregulation of SMIM3 in peripheral blood monocytes from CAD patients (Figure 6C). Collectively, these findings indicate that SMIM3 plays a potential progressive role in CAD.
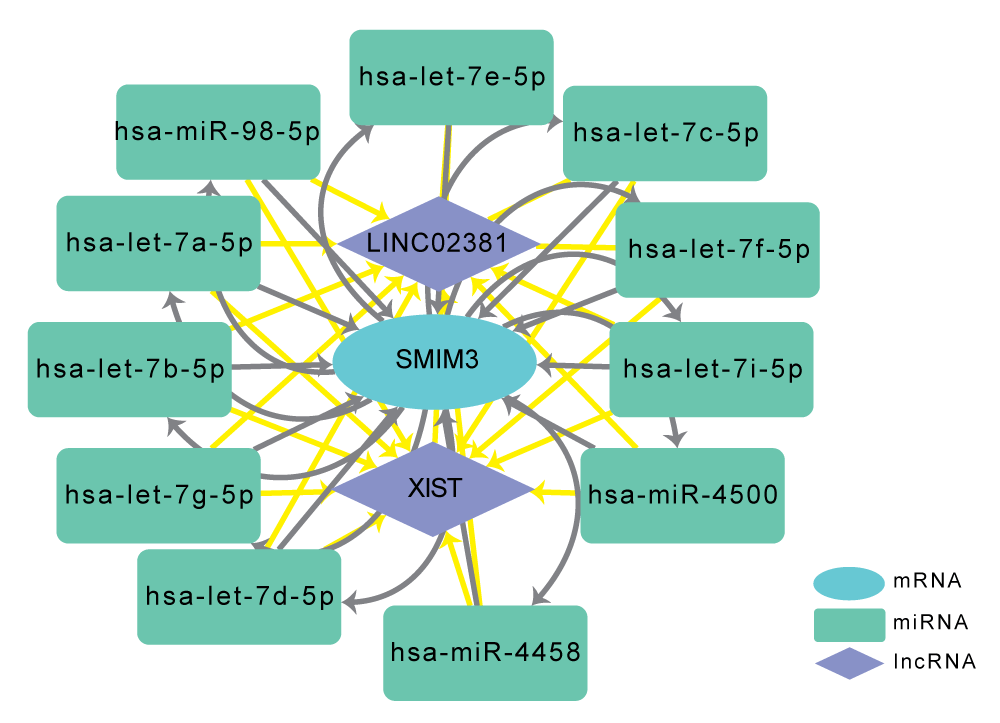
Construction of the SMIM3 regulated ceRNA network
It is well established that miRNAs can induce gene silencing and downregulate gene expression by binding to target mRNAs [26]. However, upstream molecules such as lncRNAs can modulate miRNA activity by binding to them, which in turn can result in the upregulation of gene expression [27]. MicroRNAs play important roles in many cellular and biological functions via the regulation of mRNA target translation. In the cardiovascular field, microRNAs are now acknowledged as fundamental in regulating the expression of genes that governs physiological and pathological myocardial adaptation to stress. This interaction among RNAs is referred to as a ceRNA network [28]. Using two online miRNA databases, we predicted 11 target miRNAs for SMIM3, including hsa-let-7i-5p, hsa-let-7f-5p, hsa-let-7c-5p, hsa-miR-4500, hsa-let-7e-5p, hsa-miR-98-5p, hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7g-5p, hsa-let-7d-5p and hsa-miR-4458. Then, we used the online database Starbase 3.0 [23] to predict the lncRNAs that interact with the 11 above miRNAs. Finally, we identified two target lncRNAs of the miRNAs targeting SMIM3: XIST and LINC02381. The ceRNA network was constructed based on these predicted interactions and visualized using Cytoscape (Figure 7). In summary, we constructed the ceRNA network to reveal the potential RNA regulatory pathways to further elucidate the underlying progress of CAD.
Discussion
CAD is a chronic inflammatory disease caused by the accumulation of fatty plaques within the cardiovascular system [29]. Its pathogenesis is mainly due to the interaction between endothelial cells dysfunction and cholesteryl lipoproteins in the subendothelial layer, and subsequent changes in endothelial cells permeability trigger infiltration of monocytes in the subendothelial layer, which differentiate into macrophages and release inflammatory mediators [30]. Current research focuses on plaque formation driving the pathogenesis of CAD, but inflammation, as a systemic process, plays a central role in the progression of atherosclerotic plaques. Recent studies have found that although patients with stable plaques are not in a high-risk state at the time of diagnosis, stable plaques can deteriorate rapidly and quickly reach a very high-risk state [31]. To further aid the prevention and early detection of CAD progression, it is crucial to gain a deeper understanding of the molecular mechanisms and regulatory pathways involved in rapid CAD progression. Therefore, this study focused on analyzing patients with progressive CAD and stable CAD. Using WGCNA and integrating the clinical characteristics of the samples, we identified the tan module as significantly associated with CAD progression among 15 modules. The results of KEGG and GO enrichment analysis showed that the tan module was mainly involved in platelet-related biological processes such as platelet activation, platelet degranulation, and platelet aggregation. Studies have demonstrated that platelets play an potential biological role in cardiovascular disease, both in the pathogenesis of atherosclerosis and acute thrombotic events [32]. Platelet activation pathways are mechanistically connected to monocyte infiltration in several ways. Platelets interact with monocytes via multiple mechanisms, including the binding of platelet P-selectin to monocyte PSGL-1, the release of platelet granules containing chemokines and cytokines, and the shedding of platelet-derived microvesicles. These interactions upregulate monocyte pro-inflammatory surface markers, enhance their migration capabilities, and promote a pro-coagulant phenotype. Activated platelets also release chemokines like CXCL4, which promote monocyte adhesion to endothelial cells and their transmigration into atherosclerotic lesions. Monocytes exposed to platelets exhibit increased expression of pro-inflammatory cytokines such as TNF-α, MCP-1, and IL-1β, and adopt a pro-atherosclerotic phenotype. This platelet-monocyte crosstalk is critical in driving inflammation and thrombosis in cardiovascular diseases [33,34]. In this work, we identified that genes such as NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3 may promote disease progression through platelet-monocyte crosstalk. Among these, LOC340508 and KIAA1211 are indeed less characterized in the literature, their specific functions in atherosclerosis or cardiovascular diseases remain largely unknown. Further experimental studies are required to elucidate their roles in these processes. Notably, the SMIM3 gene, encoding small integral membrane protein 3, was associated with monocyte cell infiltration, and could achieve protein binding function in monocytes probably by having structural similarity to cytokine receptor common subunit beta, as predicted by the SWISS-MODEL database (https://swissmodel.expasy.org/repository/uniprot/Q9BZL3). It suggested SMIM3 may regulate monocyte differentiation, contributing to plaque instability, thereby promoting the rapid progression of CAD.
Furthermore, antiplatelet drugs have been used to treat CAD and acute coronary syndrome, which further supports the role of platelets in CAD progression [35]. Then, we further screened genes specifically up-regulated in progressive CAD by DEA and finally obtained the CAD progression-associated genes: NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3. NID2, namely nidogen 2, encodes a cell-adhesion protein that binds to collagens I and IV, as well as laminin, potentially maintaining basement membrane structure. Previous studies had identified SNP loci of NID2 associated with the pathogenesis of CAD, but the underlying mechanism remained unclear [36]. Through GSEA, we observed that NID2 was mainly involved in coronary vasculature development, microtubule anchoring at centrosome, microtubule anchoring at microtubule organizing center, positive regulation of interleukin-13 production, and regulation of monocyte differentiation pathways. Additionally, we identified five new key genes related to CAD progression. The regulatory pathways are similar to those of NID2, all of which can regulate monocyte differentiation according to GSEA. Monocytes participate in many pathophysiological pathways of CAD, such as lipid metabolism, coagulation, apoptosis, hypoxia, angiogenesis, and immune response, playing an important role in various stages of coronary heart disease [37]. Different subtypes of monocytes exert distinct effects, including pro-inflammatory, anti-inflammatory, pro-collagen deposition, and pro-angiogenesis. The dynamic conversion of their subpopulation ratios play a crucial role in the stability of atherosclerotic plaques [38]. Using immune infiltration and correlation analysis, we found that SMIM3 was significantly positively correlated with the infiltration of monocytes, suggesting that SMIM3 was involved in the monocyte infiltration and differentiation, and potentially alter pro-inflammatory monocyte phenotypes, leading to plaque instability, thereby promoting the rapid progression of CAD.
The results differ from those of previous studies further reflects the promoting effect of platelets on the development of CAD. The possible reasons are as follows. Specifically, our approach differs from that of Nührenberg, et al. [16] in identifying CAD progression-related genes. The advantage of the WGCNA algorithm [11] we applied is that the algorithms spread thousands of genes into several co-expression modules based on their expression patterns which provides a new system biology method based on microarray or RNA-seq data and efficiently and accurately uncovers relationships between gene networks and sample traits, even for detecting low-abundance or weakly expressed genes. We would like to promote the prevention and screening of CAD progression.
Additionally, understanding the molecular mechanisms and regulatory pathways underlying rapid CAD progression may facilitate the development of small molecule targeted therapies to slow disease progression. In the cardiovascular field, microRNAs are now recognized as key regulators of gene expression that control physiological and pathological adaptations of heart muscle to stress. Their high stability, resistance to degrading enzymes, and ease of detection make them reliable non-invasive biomarkers. Here, we aim to identify microRNAs related to SMIM3 through a ceRNA network. We have successfully identified miRNAs associated with SMIM3 as well as lncRNAs that interact with them, which allows us to further elucidate potential SMIM3-related regulatory pathways. This approach will contribute to a more comprehensive understanding of the role of SMIM3 in CAD.
However, several limitations should be acknowledged when explaining those results in our study. First, the datasets used for analysis and verification are relatively small due to limited resources. A study with a large amount of data is needed to verify our results. Second, additional experiments need to be conducted to verify our analysis results. While peripheral blood gene profiling provides a minimally invasive alternative for molecular assessment [39-41], it faces significant limitations. Peripheral blood-derived gene expression profiles often fail to fully recapitulate localized molecular dynamics within coronary plaques due to the inherent heterogeneity of circulating blood cells and the unique plaque microenvironment. In contrast, direct tissue expression analysis, despite its invasive nature, offers superior resolution in characterizing plaque biology by capturing cell type-specific transcriptional signatures shaped by localized pathological processes, including inflammatory cascades, hypoxia, and oxidative stress. The weak correlation between peripheral blood biomarkers and plaque pathology arises from confounding systemic factors, such as metabolic fluctuations, comorbid conditions, and genetic variability, that independently modulate circulating transcriptional patterns. Conversely, tissue-based profiling enables the discovery of plaque-specific therapeutic targets and instability-associated biomarkers that remain obscured in peripheral studies. Nevertheless, the clinical risks and logistical challenges of repetitively acquiring coronary plaque specimens restrict its utility in longitudinal or population-scale research. Therefore, striking a balance between these approaches is crucial, underscoring the identification of noninvasive and effective molecular markers as a key direction for future research.
Conclusion
Our work identified six key genes in the crucial module, NID2, PGRMC1, TSC22D1, LOC340508, KIAA1211, and SMIM3, as potential biomarkers for the diagnosis and treatment of progressive CAD, and SMIM3 expression in monocytes may contribute to regulate disease progression in CAD.
Author contributions
GZW and ZJR designed and performed the work. HXC, YC, MXK, and JZC contributed to the acquisition and analysis of the work. LS and HG designed the work. SHL contributed to the interpretation of the work. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Jiangsu Association for Science & Technology Youth Science & Technology Talents Lifting Project (Grant No. JSTJ-2023-XH015).
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Availability of data and materials
The data sets used in the present research were described in the Data collection and analysis section. The code used in our study can be obtained from the corresponding author.
Ethics approval and consent to participate
This study involves human participants and was approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang University (I20211258(1)). We confirmed that informed consent was obtained from all subjects and/or their legal guardian(s). All experiments were performed in accordance with relevant guidelines and regulations.
The authors would like to thank the Department of Vascular Surgery of the Second Affiliated Hospital of Zhejiang University for their technical support and guidance during the study.
- McCullough PA. Coronary artery disease. Clin J Am Soc Nephrol. 2007;2(3):611-6. Available from: https://doi.org/10.2215/cjn.03871106
- Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111(25):3481-8. Available from: https://doi.org/10.1161/circulationaha.105.537878
- Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114(12):1852-66. Available from: https://doi.org/10.1161/circresaha.114.302721
- Popa LE, Petresc B, Cătană C, Moldovanu CG, Feier DS, Lebovici A, et al. Association between cardiovascular risk factors and coronary artery disease assessed using CAD-RADS classification: a cross-sectional study in Romanian population. BMJ Open. 2020;10(2):e031799. Available from: https://doi.org/10.1136/bmjopen-2019-031799
- Head SJ, Milojevic M, Daemen J, Ahn JM, Boersma E, Christiansen EH, et al. Mortality after coronary artery bypass grafting versus percutaneous coronary intervention with stenting for coronary artery disease: a pooled analysis of individual patient data. Lancet. 2018;391(10124):939-48. Available from: https://doi.org/10.1016/s0140-6736(18)30423-9
- Kadota A. The estimated absolute risk of coronary artery disease and subclinical atherosclerosis. J Atheroscler Thromb. 2021;28(12):1260-2. Available from: https://doi.org/10.5551/jat.ED177
- Badimon L, Padró T, Vilahur G. Atherosclerosis, platelets and thrombosis in acute ischaemic heart disease. Eur Heart J Acute Cardiovasc Care. 2012;1(1):60-74. Available from: https://doi.org/10.1177/2048872612441582
- Björkegren JLM, Lusis AJ. Atherosclerosis: recent developments. Cell. 2022;185(10):1630-45. Available from: https://doi.org/10.1016/j.cell.2022.04.004
- Pasalic L, Wang SS, Chen VM. Platelets as biomarkers of coronary artery disease. Semin Thromb Hemost. 2016;42(3):223-33. Available from: https://doi.org/10.1055/s-0036-1572328
- Mori H, Torii S, Kutyna M, Sakamoto A, Finn AV, Virmani R. Coronary artery calcification and its progression: what does it really mean? JACC Cardiovasc Imaging. 2018;11(1):127-42. Available from: https://doi.org/10.1016/j.jcmg.2017.10.012
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. Available from: https://doi.org/10.1186/1471-2105-9-559
- Qi B, Chen JH, Tao L, Zhu CM, Wang Y, Deng GX, et al. Integrated weighted gene co-expression network analysis identified that TLR2 and CD40 are related to coronary artery disease. Front Genet. 2020;11:613744. Available from: https://doi.org/10.3389/fgene.2020.613744
- Wang Y, Liu T, Liu Y, Chen J, Xin B, Wu M, Cui W. Coronary artery disease associated specific modules and feature genes revealed by integrative methods of WGCNA, MetaDE and machine learning. Gene. 2019;710:122-30. Available from: https://doi.org/10.1016/j.gene.2019.05.010
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-50. Available from: https://doi.org/10.1073/pnas.0506580102
- Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453-7. Available from: https://doi.org/10.1038/nmeth.3337
- Nührenberg TG, Langwieser N, Binder H, Kurz T, Stratz C, Kienzle RP, et al. Transcriptome analysis in patients with progressive coronary artery disease: identification of differential gene expression in peripheral blood. J Cardiovasc Transl Res. 2013;6(1):81-93. Available from: https://doi.org/10.1007/s12265-012-9420-5
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. Available from: https://doi.org/10.1093/nar/gkv007
- Wang C, Song C, Liu Q, Zhang R, Fu R, Wang H, et al. Gene expression analysis suggests immunological changes of peripheral blood monocytes in the progression of patients with coronary artery disease. Front Genet. 2021;12:641117. Available from: https://doi.org/10.3389/fgene.2021.641117
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284-7. Available from: https://doi.org/10.1089/omi.2011.0118
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. Available from: https://doi.org/10.1186/1471-2105-12-77
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005. Available from: https://doi.org/10.7554/elife.05005
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127-31. Available from: https://doi.org/10.1093/nar/gkz757
- Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92-7. Available from: https://doi.org/10.1093/nar/gkt1248
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-504. Available from: https://doi.org/10.1101/gr.1239303
- Tousoulis D, Paroutoglou IP, Papageorgiou N, Charakida M, Stefanadis C. Recent therapeutic approaches to platelet activation in coronary artery disease. Pharmacol Ther. 2010;127(2):108-20. Available from: https://doi.org/10.1016/j.pharmthera.2010.05.001
- Ali Sheikh MS, Alduraywish A, Almaeen A, Alruwali M, Alruwaili R, Alomair BM, et al. Therapeutic value of miRNAs in coronary artery disease. Oxid Med Cell Longev. 2021;2021:8853748. Available from: https://doi.org/10.1155/2021/8853748
- Ebadi N, Ghafouri-Fard S, Taheri M, Arsang-Jang S, Parsa SA, Omrani MD. Dysregulation of autophagy-related lncRNAs in peripheral blood of coronary artery disease patients. Eur J Pharmacol. 2020;867:172852. Available from: https://doi.org/10.1016/j.ejphar.2019.172852
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344-52. Available from: https://doi.org/10.1038/nature12986
- Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868-74. Available from: https://doi.org/10.1038/nature01323
- Libby P, Lee RT. Matrix matters. Circulation. 2000;102(16):1874-6. Available from: https://doi.org/10.1161/01.cir.102.16.1874
- Fox KAA, Metra M, Morais J, Atar D. The myth of 'stable' coronary artery disease. Nat Rev Cardiol. 2020;17(1):9-21. Available from: https://doi.org/10.1038/s41569-019-0233-y
- Schrör K, Huber K. Platelets, inflammation and anti-inflammatory drugs in ACS and CAD. Thromb Haemost. 2015;114(3):446-8. Available from: https://doi.org/10.1160/th15-08-0632
- Rolling CC, Barrett TJ, Berger JS. Platelet-monocyte aggregates: molecular mediators of thromboinflammation. Front Cardiovasc Med. 2023;10:960398. Available from: https://doi.org/10.3389/fcvm.2023.960398
- Magrone T, Magrone M, Russo MA, Jirillo E. Platelets: angels and demons dancing on the immune stage. Nutrition conducts the orchestra. Endocr Metab Immune Disord Drug Targets. 2021;21(7):1196-1218. Available from: http://dx.doi.org/10.2174/1871530320666200901183119
- Müller KA, Chatterjee M, Rath D, Geisler T. Platelets, inflammation and anti-inflammatory effects of antiplatelet drugs in ACS and CAD. Thromb Haemost. 2015;114(3):498-518. Available from: http://dx.doi.org/10.2174/1871530320666200901183119
- Abramowitz Y, Roth A, Keren G, Isakov O, Shomron N, Laitman Y, et al. Whole-exome sequencing in individuals with multiple cardiovascular risk factors and normal coronary arteries. Coron Artery Dis. 2016;27(4):257-66. Available from: https://doi.org/10.1097/mca.0000000000000357
- Ghattas A, Griffiths HR, Devitt A, Lip GY, Shantsila E. Monocytes in coronary artery disease and atherosclerosis: where are we now? J Am Coll Cardiol. 2013;62(17):1541-51. Available from: https://doi.org/10.1016/j.jacc.2013.07.043
- Pamukcu B, Lip GY, Devitt A, Griffiths H, Shantsila E. The role of monocytes in atherosclerotic coronary artery disease. Ann Med. 2010;42(6):394-403. Available from: https://doi.org/10.3109/07853890.2010.497767
- Vanderlaan PA, Reardon CA. Thematic review series: the immune system and atherogenesis. The unusual suspects: an overview of the minor leukocyte populations in atherosclerosis. J Lipid Res. 2005;46(5):829-38. Available from: https://doi.org/10.1194/jlr.r500003-jlr200
- Patino WD, Mian OY, Kang JG, Matoba S, Bartlett LD, Holbrook B, et al. Circulating transcriptome reveals markers of atherosclerosis. Proc Natl Acad Sci U S A. 2005;102(9):3423-8. Available from: https://doi.org/10.1073/pnas.0408032102
- Tabibiazar R, Wagner RA, Deng A, Tsao PS, Quertermous T. Proteomic profiles of serum inflammatory markers accurately predict atherosclerosis in mice. Physiol Genomics. 2006;25(2):194-202. Available from: https://doi.org/10.1152/physiolgenomics.00240.2005